 |
|
Study Design
Data Insights
Additional Insights
Supplementary Materials Download: |
This site supports Harbison et al. The work is a collaboration between the laboratories of Rick Young and Ernest Fraenkel. DNA-binding transcriptional regulators interpret the genome’s regulatory code by binding to specific sequences to induce or repress gene expression. Comparative genomics has recently been used to identify potential cis-regulatory sequences within the yeast genome on the basis of phylogenetic conservation, but this information alone does not reveal if or when transcriptional regulators occupy these binding sites. We have constructed a draft of yeast’s transcriptional regulatory code by mapping the sequence elements that are bound by regulators under various conditions and that are conserved among Saccharomyces species. The organization of regulatory elements in promoters and the environment-dependent use of these elements by regulators are discussed. We find that environment-specific use of regulatory elements predicts mechanistic models for the function of a large population of yeast’s transcriptional regulators. 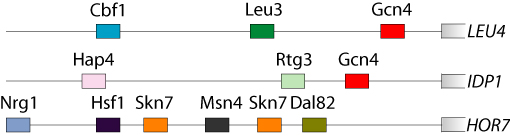 |
| YOUNG
LAB
Whitehead Institute 9 Cambridge Center Cambridge, MA 02142 [T] 617.258.5218 [F] 617.258.0376 Contact us |