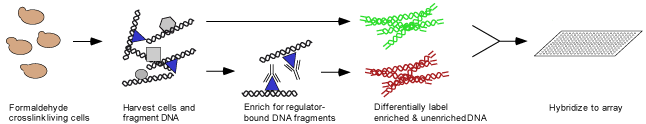
Technology - Location Analysis
Genome-wide location analysis was performed as previously described (Ren et al., 2000). Bound proteins were formaldehyde-crosslinked to DNA in vivo, followed by cell lysis and sonication to shear DNA. Crosslinked material was immunoprecipitated with an anti-myc antibody, followed by reversal of the crosslinks to separate DNA from protein (Aparicio, 1999; Orlando et al., 2000). Immunoprecipitated DNA and DNA from an unenriched sample were amplified and differentially fluorescently labeled by ligation-mediated PCR. These samples were hybridized to a microarray consisting of spotted PCR products representing the intergenic regions of the S.cerevisiae genome.

Microarray design
Using the Yeast Intergenic Region Primer set (Research Genetics) we PCR amplified
and printed on the order of 6000 spots, representing essentially all of the
known intergenic regions in the yeast genome. The average size of the spotted
PCR products was 480 bp, and the sizes ranged from 60 bp to 1500 bp. Details
of how the microarray was produced can be found here.