A Chromatin Landmark and Transcription Initiation at
Most Promoters in Human Cells
Data
Figures
Supplemental Information
Acknowledgements
References
|
Figure 4
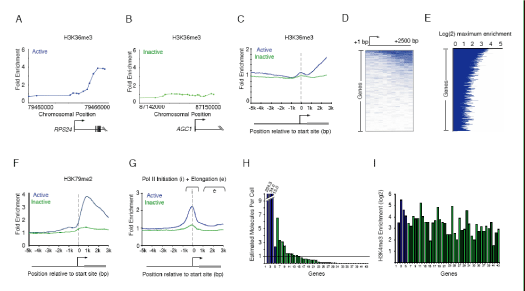
Figure 4. Events associated with transcriptional elongation occur at a
minority of genes.
A-B. Enrichment ratios are shown for H3K36me3, which is associated with
elongation. The start and direction of transcription are noted by arrow.
C. Composite enrichment profiles for H3K36me3 at genes called present or
absent for mRNA transcripts using microarray data (as in Figure 2G). The start
and direction of transcription of the average gene is noted by arrow.
D. Single probe enrichment ratios of H3K36me3 across the average 2.5kb region
downstream of the transcription start site (10 probes per promoter). Enriched
probes (blue) and unenriched probes (white) are shown for each gene. Individual
genes are represented top to bottom and ordered by average binding ratio
across the entire region.
E. H3K36me3 maximum enrichment (blue horizontal line) between 1kb and 3kb
of the transcription start site for each gene represented on human promoter
array. Genes are ordered as in (D).
F. Composite enrichment profiles for H3K79me2 at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G).
G. Composite enrichment profiles for Pol II (4H8) at genes called present or absent for mRNA transcripts using microarray data (as in Figure 2G). Both the initiation and elongating forms or Pol II are recognized by this antibody. The area from 1kb to 3kb downstream of the promoter (designated e) was used to determine genes enriched. The start and direction of transcription of the average gene is noted by arrow.
H. Quantitative RT-PCR analysis of total RNA extracted from hES cells. Estimated transcripts per cell were obtained by dividing the number of molecules of cDNA, as determined by relative quantitation using the standard curve in Figure S5 by the number of cells used for the amplification. Positive control probes (active genes, blue bars) and test sample probes (Inactive genes, green bars) are as follows: (1) No probe (2)RPLPO, (3)HMGB2, (4)CCNB1, (5)EIF2C3, (6)HOP, (7)GUCY1A3, (8)KITLG, (9)SOX9, (10)PLD1, (11)INCENP, (12)ERCC4, (13)CSF1, (14)HOXB1, (15)GPR143, (16)DSC3, (17)NFKBIL2, (18)ANG, (19)LEFTY1, (20)LNPEP, (21)TCF21, (22)RAP1GA1, (23)KLF2, (24)KCNJ3, (25)TNFSF7, (26)PRRG2, (27)FOXG1B, (28)DLX5, (29)KCNH1, (30)ACADL, (31)CXCL2, (32)HOXD12, (33)ONECUT1, (34)LYST, (35)PCSK2, (36)AVPR1B, (37)SMP3, (38)EPHX2, (39)UCP1, (40)AGC1, (41)ERAF, (42)HBA1, (43)MLNR). Measured values for the genes can be found in Table S6. Broken axis is indicated by white line.
I. H3K4me3 maximum enrichment (within 1kb of the transcription start site) for
each gene represented in (H).
|

