A Chromatin Landmark and Transcription Initiation at
Most Promoters in Human Cells
Data
Figures
Supplemental Information
Acknowledgements
References
|
Figure 1
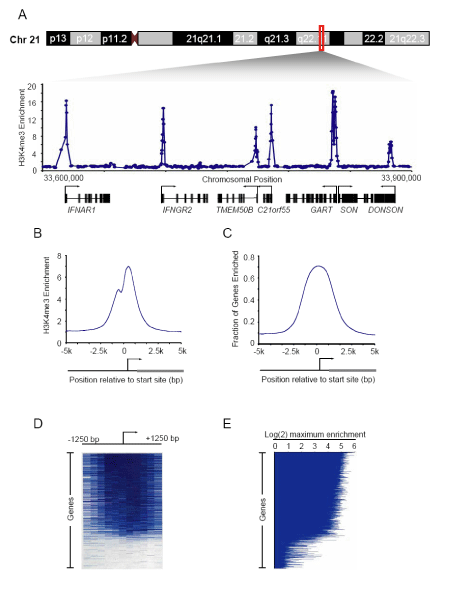
Figure 1. H3K4me3 modified nucleosomes are found at sites of
transcription initiation.
A. Genomic DNA segments enriched for H3K4me3 modified histones in hES
cells were identified with ChIP-Chip using DNA microarrays tiling the non-repeat
human genome. Enrichment ratios for H3K4me3 across 300kb of chromosome
21 are shown. Start sites and direction of transcription are indicated by arrows. See Supplemental Table S1 for the regions enriched for H3K4me3 on chromosome 21.
B. Composite H3K4me3 enrichment profile for all known genes. The start and
direction of transcription of the average gene is noted by arrow. Details on
composite profile generation are described in supplemental data.
C. Fraction of genes enriched for H3K4me3 (enrichment ratio >2) was calculated
for all known promoters. All genes were aligned according to the position of their
transcription start sites and the fraction of genes with enriched H3K4me3 binding
was then calculated in a sliding 200 bp window.
D. Single probe enrichment ratios of H3K4me3 across the 2.5kb region
surrounding the transcription start site (~10 probes per promoter region).
Enriched probes (blue) and unenriched probes (white) are shown for each gene.
Individual promoters are represented top to bottom and ordered by average
binding ratio within a 1kb window surrounding the transcription start site.
E. H3K4me3 maximum enrichment (blue horizontal line) within 1kb of the
transcription start site for each gene represented on the whole genome array.
Genes are ordered as in (D).
|

