A Chromatin Landmark and Transcription Initiation at
Most Promoters in Human Cells
Data
Figures
Supplemental Information
Acknowledgements
References
|
Figure 2
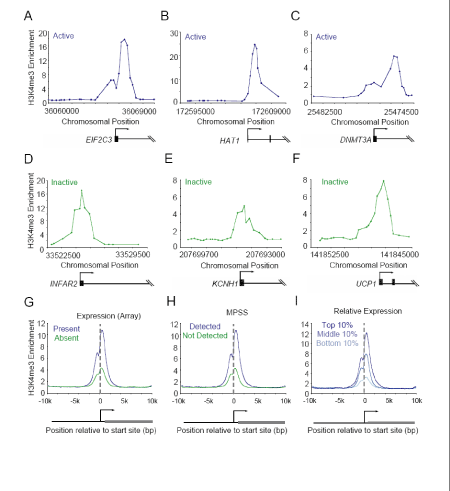
Figure 2. H3K4me3 modified nucleosomes are enriched at both active and
inactive promoters.
A-F. Genomic DNA segments enriched for H3K4me3 modified histones were
identified with ChIP-Chip using DNA microarrays spanning the entire non-repeat portion of the human genome. Enrichment ratios are shown for H3K4me3 at
active genes (mRNA transcript called present in Affymetrix expression data and
detected by Massively Parallel Signature Sequencing (MPSS) data) and inactive
genes (mRNA transcript called absent in Affymetrix expression data and not
detected by MPSS data). Gene start and direction of transcription are indicated
by arrows.
G-I. Composite H3K4me3 enrichment profiles for G) genes called present or
absent for mRNA transcript using Affymetrix data (Sato et al., 2003; Abeyta et al.,
2004), H) genes detected or not detected by MPSS (Brandenberger et al., 2004;
Wei et al., 2005) and I) genes in the top 10%, middle 10%, or bottom 10% by
relative expression (Su et al., 2004).
|

