 |
|
Activated Signal Transduction Kinases Frequently Occupy Target Genes
Acknowledgements |
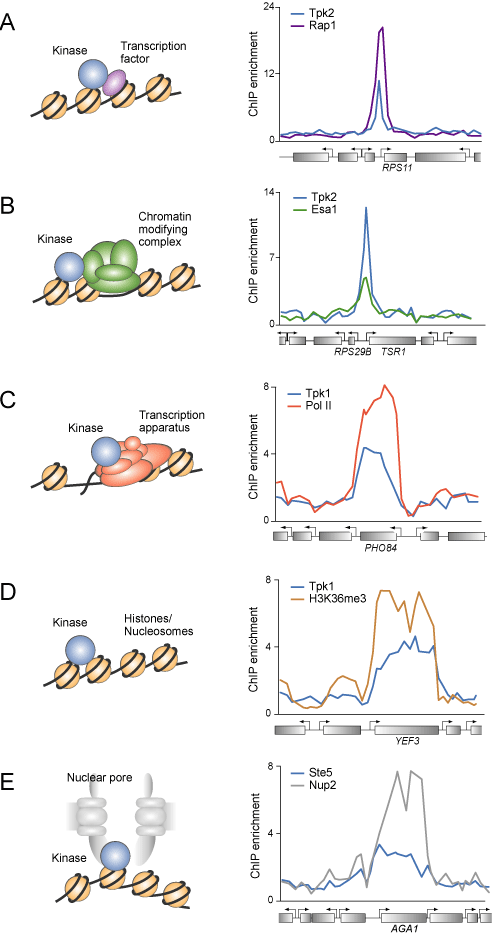
Signal transduction kinases may occupy DNA through interaction with (A) transcriptional regulators (purple), (B) chromatin regulators (green), (C) transcription apparatus (red), (D) nucleosomes (brown) and (E) nuclear pore proteins (grey). Signal transduction pathways can regulate transcription through phosphorylation of transcription factors, components of chromatin modifying complexes, the transcription apparatus, and histones. In most instances, the kinases were thought to interact transiently with their substrates rather than to occupy transcription factors and other substrates at the genes they regulate. The model described here is that signaling kinases in many cases are directly associated with target genes, either by binding directly to protein complexes associated with their substrates or through adaptor proteins. Association with actively transcribed genes may also occur through interaction with nuclear pore proteins, to which actively transcribed genes may be recruited. Comparing the pattern of gene occupancy of kinases with those of other proteins detected by ChIP-chip suggests models by which kinases may associate with DNA. Experimental examples for possible co-occupancy are shown to the right: (A) Tpk2p and the DNA-binding transcription factor Rap1p. (B) Tpk2p and the Esa1p subunit of the chromatin modifying NuA4 complex. (C) Tpk1p and RNA Polymerase II of the transcription apparatus. (D) Tpk1p and the histone H3K36-trimethyl modification, which is deposited during transcription. (E) Ste5p and the nuclear pore protein Nup2p, which is part of the nuclear pore complex. |
| YOUNG
LAB
Whitehead Institute 9 Cambridge Center Cambridge, MA 02142 [T] 617.258.5218 [F] 617.258.0376 CONTACT US |