|
|
|
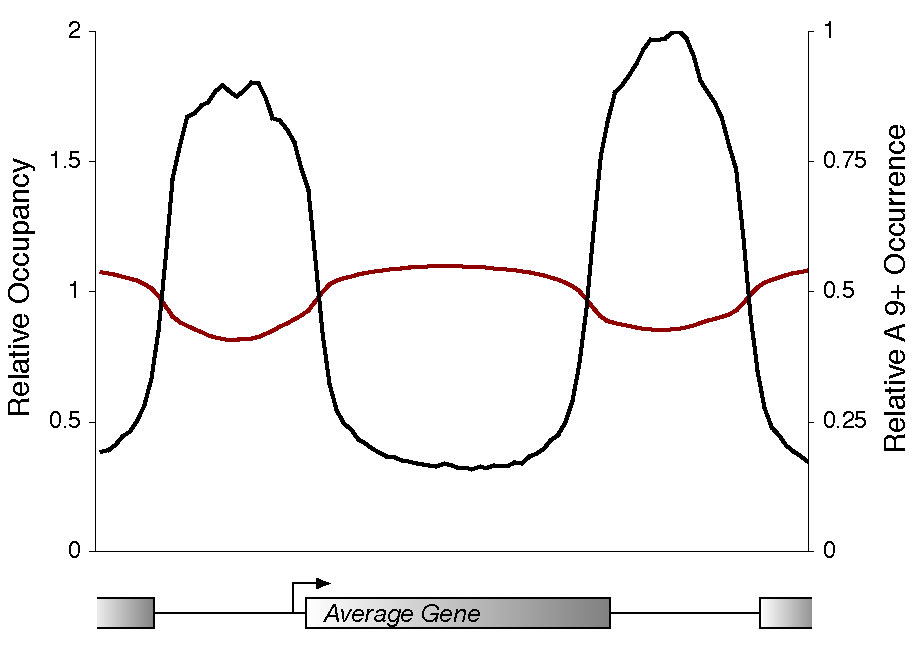
Composite profile of histone occupancy plotted against relative poly A9 occurrence. Probes corresponding to all genes were weighted according to the number of nearby A9 (or T9) tracts. The resulting occupancy is relative to the maximum average occupancy. |
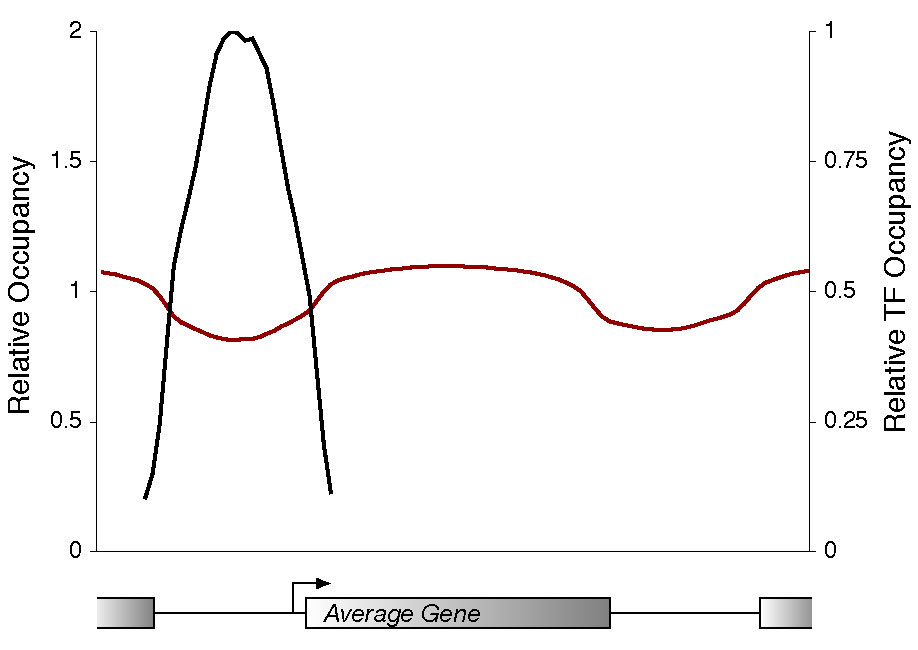
Relative occupancy of intergenic regions bound by transcriptional regulators (P <0.001) and containing matches to cognate consensus binding specificities determined in Harbison et al (2004). Probes corresponding to all promoter regions were weighted according to the number of nearby regulator-bound binding sites. The resulting occupancy is relative to the maximum average occupancy. |
| To explore whether poly(dA:dT) sequences are associated with reduced nucleosome occupancy, we examined the frequency of their association with nucleosome-depleted promoter regions. We found that the distribution of poly A9 sites (in black, above) is strongly anti-correlated with nucleosome occupancy (in red, above), peaking within intergenic regions and at the lowest levels throughout open reading frames. Approximately 77% of poly A9 sites located within promoter regions coincide with sites of minimal nucleosome occupancy. Furthermore, unlike binding sites for transcriptional regulators, which tend to occur upstream of genic regions, poly A9 elements occur frequently within downstream regions. This could explain why histone minima occur frequently in intergenic regions that are downstream of two convergently transcribed genes. | We investigated whether the binding sites occupied by most transcriptional regulators (Harbison et al., 2004 in black, above) are associated with minimal nucleosome occupancy (in red, above). Of the nearly 2000 promoter regions that are occupied at least by one transcriptional regulator at high confidence (P value less than 0.001), 74% showed minimal nucleosome occupancy in the vicinity of the transcription factor binding site. Furthermore, we noted an association between promoter binding by multiple regulators and reduced nucleosome occupancy. In agreement with a previous study (Bernstein et al., 2004), we note that certain transcription factors (Rap1, Fhl1, Swi4) were more likely to have strong associations with nucleosome minima (up to a 90% overlap) than others. |
These results strongly indicate that certain DNA sequences are associated with regions of low nucleosome occupancy, although they do not show that the presence of these sequences themselves are responsible for altered nucleosome occupancy. In order to test this hypothesis, we created 5 strains in which both poly(dA:dT) and transcription factor binding sites had been removed singly or in combination. Surprisingly, deletion of these sites failed to confer any changes on the relative occupancy of nucleosomes, indicating that other sequences or mechanisms must contribute to the control of nucleosome density. |
|
.