High Resolution ChIP-chip
In an effort
to overcome the limitations to understanding global chromatin structure imposed
by standard microarray formats, we designed a yeast microarray with more than
40,000 probes, positioned at an average density of 266 bp. An example of the
results of combining these arrays with chromatin immunoprecipitation of an epitope-tagged
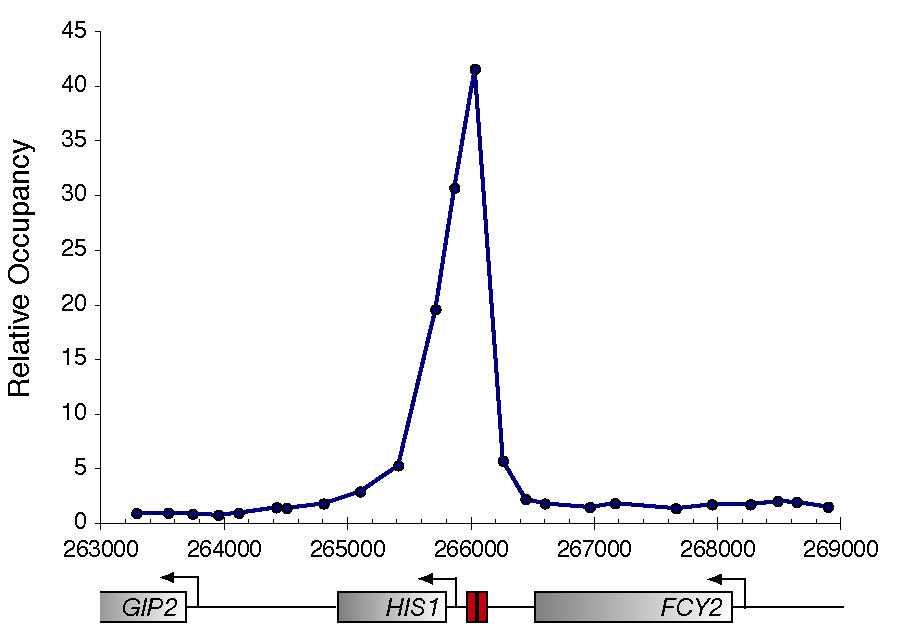
protein DNA-binding protein (Gcn4) is shown below. A sequence match to the Gcn4
binding specificity is depicted as a red box. On the left is an example profile
of Gcn4 enrichement across a region known to be bound by Gcn4. At right is a
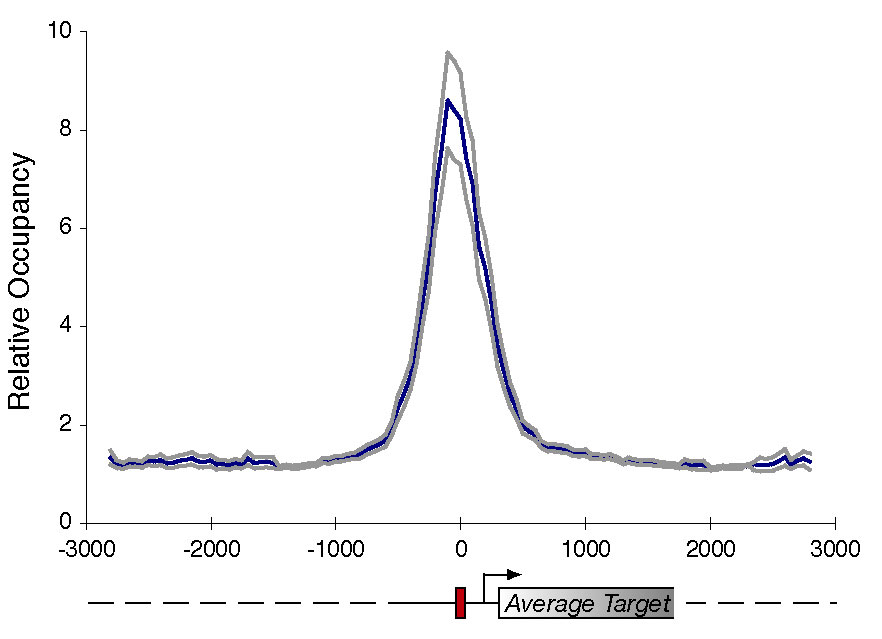
composite of 84
such regions. More examples are shown here.
Based on the analysis of Gcn4 targets, we estimate
a false positive rate of less than 1% and a false negative rate of ~25%, corresponding
with a total of 210 genes whose promoters are bound within the optimal P
value threshold of 6x10-6.